Biological Science 2024/02/07
Researchers from NAIST uncover the cellular process governing petal abscission, revealing the crucial role of jasmonic acid and autophagy.
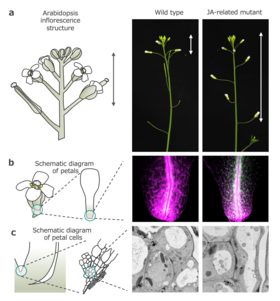
Organogenesis, an important aspect of flowering, helps in understanding key processes of plant development like the formation of floral organs, attainment of reproductive capability, and abscission leading to seed and fruit development. Although abscission, a physiological process involving the shedding of plant organs from the main plant body, may seem opposite to the conventional definition of development, it significantly impacts plant reproductive success and seed dispersal in angiosperms. Furthermore, the petal abscission is dependent on new RNA and protein synthesis, along with cell-wall collapse and cytoplasmic and vacuolar reduction, suggesting that, instead of being a mere catastrophic event, it is an actively controlled cellular process.
Notably, the petal base cells in various plants, like the Japanese morning glory, exhibit distinctive changes in vesicle numbers and cytoplasmic components, hinting at the role of a process called "autophagy" (an intracellular degradation process that allows cells to recycle damaged intracellular components) in petal abscission. However, how these cellular changes are spatiotemporally regulated and lead to autophagy has remained a persistent challenge.
To address this, a team of researchers led by Nobutoshi Yamaguchi and Toshiro Ito from Nara Institute of Science and Technology in Japan used Arabidopsis thaliana Col-0 and mutants/transgenic lines. The study, published in the journal Nature Communications , surpasses conventional boundaries, and explores autophagy by utilizing advanced proximity ligation assays (PLA) technology. The researchers employed various experimental methodologies and analyses to investigate the intricate mechanisms governing petal abscission, quantify petal abscission timings, and examine the position of flowers when petals detached. Speaking about the findings, Yamaguchi says, "Our work describes a phytohormone-mediated chromatin state switch that controls spatiotemporal-specific activation of autophagy, leading to terminal cell differentiation for petal abscission."
The researchers found that AGAMOUS, the gene responsible for specifying stamen and carpel identity, and jasmonic acid (JA) promote petal abscission at petal bases through cellular differentiation. They also identified a JA-regulated chromatin state switch at the base of petals that directed local cell fate determination via autophagy.
They discovered that during petal maintenance, the co-repressors of JA signaling assemble at the base of petals to inhibit MYC activity, which results in lower levels of ROS. But when JA accumulates at the petal bases, it triggers chromatin remodeling, allowing MYC factors to promote chromatin accessibility for their downstream targets. Of these targets, ANAC102 specifically accumulates before abscission to increase ROS levels and induce ATG, thus triggering autophagy. The induced autophagy at the petal base regulates maturation, vacuolar delivery, and breakdown of autophagosomes for terminal cell differentiation.
In conclusion, the study brings forth a crucial understanding of the mechanism of petal senescence, highlighting the regulatory role of the JA pathway in orchestrating autophagosome maturation and breakdown.
The insights of this study extend beyond the subject of petal abscission, focusing on development, aging, and responses to environmental cues. Yamaguchi explains "The insights gained from our study could pave the way for enhanced predictability and manipulation of the timing of petal abscission in ornamental plants. This flexibility and reversibility in controlling the shedding of petals hold promise for advancements in horticulture and agriculture."
###
Resource
- Title: Petal abscission is promoted by jasmonic acid-induced autophagy at Arabidopsis petal bases
- Authors: Yuki Furuta, Haruka Yamamoto, Takeshi Hirakawa, Akira Uemura, Margaret Anne Pelayo, Hideaki Iimura, Naoya Katagiri, Noriko Takeda-Kamiya, Kie Kumaishi, Makoto Shirakawa, Sumie Ishiguro, Yasunori Ichihashi, Takamasa Suzuki, Tatsuaki Goh, Kiminori Toyooka, Toshiro Ito, Nobutoshi Yamaguchi
- Journal: Nature Communications
- DOI: 10.1038/s41467-024-45371-3